aGrUM/pyAgrum 1.0.0 and fast inferences
Posted on Fri 22 April 2022 in News
The 1.0.0 version of aGrUM is finally here and brings its share of new features. The most important is the parallelization of the inference methods that is the subject of this article. In particular, we will present a benchmark between the parallelized version and the old version.
But first, let's remind the exact algorithms available in aGrUM to perform inferences. There are three of them: VariableElimination, ShaferShenoy and LazyPropagation. For our comparisons, we will restrict ourselves to the latter [Madsen, A. L., & Jensen, F. V. (1999)].
To use it, let us first load the ASIA network which, like all the structures that will be used in the following, is available here.
1 2 | import pyAgrum as gum asia_bn = gum.loadBN("asia.bif") |
Now we can make an inference to determine the probability that a patient has lung cancer knowing that he has dyspnoea. To do this we need to create an inference engine and call the makeInference() method:
3 4 | ie = gum.LazyPropagation(asia_bn) ie.makeInference() |
To obtain the wanted probability, we now need to specify the evidence and call the posterior() method:
5 6 | ie.addEvidence('dyspnoea', 'yes') p = ie.posterior('lung_cancer') |
In this case, printing the variable p gives us:
lung_cancer |
yes |no |
---------|---------|
0.1028 | 0.8972 |
For more examples, you can consult the notebooks about inference.
I wanna make a supersonic lemon out of you
The makeInference() method carries out the main calculation to obtain the probability of interest. Therefore, we chose to use its execution time as a metric for our comparisons between single and multi-threaded versions. We can check the number of threads that are used for the inference using getNumberOfThreads and it can be modified using setNumberOfThreads():
7 8 | print(ie.getNumberOfThreads()) ie.setNumberOfThreads(10) |
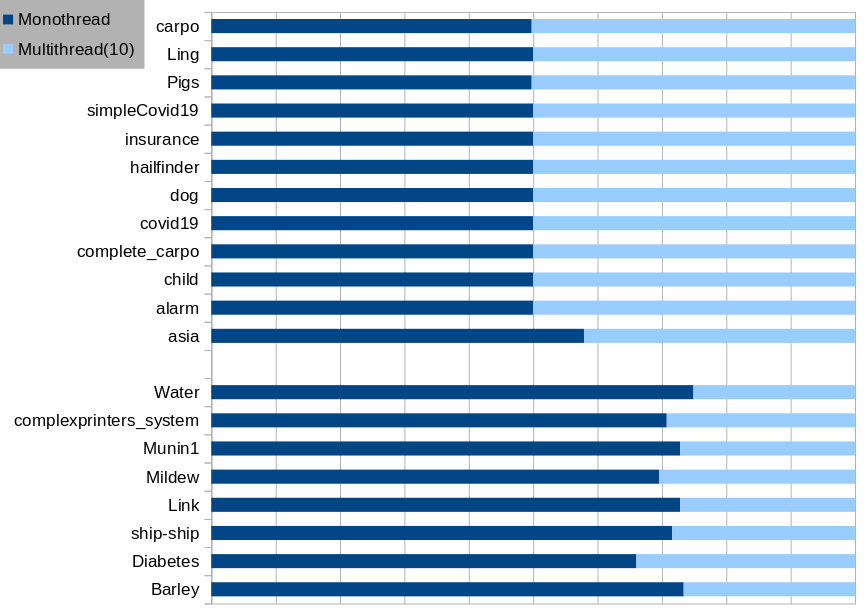
For our experiments we used successively n=1 and n=10 threads to make inferences on the different structures that can be found in the previous linked repository. For a given structure, we ran 20 iterations and the following table reports the average time of each algorithm. These experiments were done using an Intel(R) Xeon(R) Silver 4116 CPU @ 2.10GHz (48 cores) and 32GB of memory.
| Structure | Nb. of nodes | Nb. of arcs | Mono-thread (ms) | Multi-thread (ms) |
|---|---|---|---|---|
| Barley | 47 | 82 | 1,124.7 | 407.9 |
| Diabetes | 413 | 602 | 245.9 | 126.5 |
| Ling | 13 | 17 | 1.4 | 1.4 |
| Link | 724 | 1125 | 88,995.1 | 33,152.1 |
| Mildew | 35 | 46 | 258.8 | 113.1 |
| Munin1 | 186 | 273 | 10,855.5 | 4,043.3 |
| Pigs | 441 | 592 | 121.7 | 122.8 |
| Water | 32 | 66 | 327.3 | 109.6 |
| alarm | 37 | 46 | 5.5 | 5.5 |
| asia | 8 | 8 | 1.1 | 0.8 |
| carpo | 61 | 74 | 11.0 | 11.1 |
| child | 20 | 25 | 2.9 | 2.9 |
| complete_carpo | 61 | 74 | 11.0 | 11.0 |
| complexprinters_system | 164 | 215 | 9,155.6 | 3,783.7 |
| covid19 | 39 | 46 | 5.3 | 5.3 |
| dog | 5 | 4 | 0.4 | 0.4 |
| hailfinder | 56 | 66 | 9.8 | 9.8 |
| insurance | 27 | 52 | 7.5 | 7.5 |
| simpleCovid19 | 8 | 7 | 0.6 | 0.6 |
| ship-ship | 50 | 75 | 46,329.7 | 18,380.0 |
As we can see, the mono-threaded version does as well as the multi-threaded version on small structures but we really save time on large structures !